Protein Structural Analysis & Design Laboratory
Department of Biochemistry & Molecular Biology
Michigan State University
East Lansing, MI 48824-1319

About Our Work
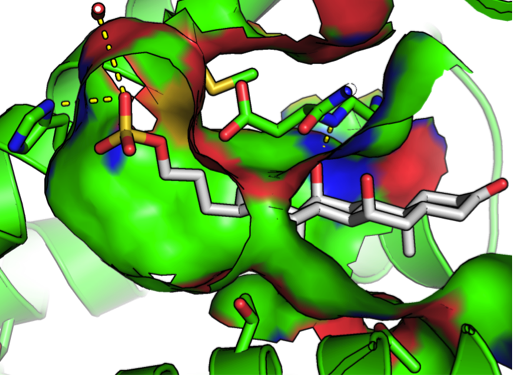
By modeling and analyzing the structures of proteins and the small organic molecules they bind, our goal is to understand how 3-dimensional molecular structures encode the preference to bind a specific partner molecule necessary for cellular function. Contributions of molecular flexibility and water molecules during binding make the process more complex and fascinating - like a molecular dance choreographed by chemistry.
A related problem we tackle is to discover the natural binding partner(s) for proteins by screening millions of molecules as potential partners, modeling their 3-dimensional interactions, and using pattern recognition to discover features shared with other protein complexes. In collaboration with experimentalists, we put our knowledge to the test through protein design and activity assays of small molecules, to address pressing environmental and medical challenges.
Leslie Kuhn, Lab Director
Professor Emeritus